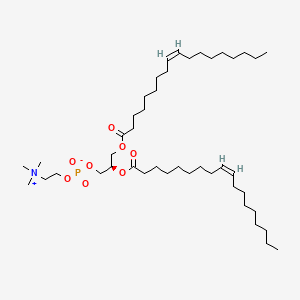
1,2-Dioleoyl-sn-Glycero-3-Phosphocholine
1,2-Dioleoyl-sn-Glycero-3-Phosphocholine is a lipid of Glycerophospholipids (GP) class. 1,2-dioleoyl-sn-glycero-3-phosphocholine is associated with abnormalities such as Exanthema, Renal tubular disorder, Nodule, Gigantism and Mycoses. The involved functions are known as Lysis, Encapsulation, Process, Uptake and Flow or discharge. 1,2-dioleoyl-sn-glycero-3-phosphocholine often locates in Cytoplasmic matrix, Endosomes, soluble, Endoplasmic Reticulum and Membrane. The associated genes with 1,2-Dioleoyl-sn-Glycero-3-Phosphocholine are P4HTM gene, synthetic peptide, BCAR1 gene, PCNA gene and CNTNAP1 gene. The related lipids are Liposomes, 1,2-oleoylphosphatidylcholine, 1,2-distearoylphosphatidylethanolamine, Butanols and Cardiolipins. The related experimental models are Mouse Model and Xenograft Model.
References related to locations published in Biochim. Biophys. Acta
| PMID | Journal | Published Date | Author | Title |
|---|---|---|---|---|
| 25687972 | Biochim. Biophys. Acta | 2015 | Evans KO et al. | Hydroxytyrosol and tyrosol esters partitioning into, location within, and effect on DOPC liposome bilayer behavior. |
| 27117642 | Biochim. Biophys. Acta | 2016 | Jurak M and Miñones J | Interactions of lauryl gallate with phospholipid components of biological membranes. |
| 26657692 | Biochim. Biophys. Acta | 2016 | Capponi S et al. | Interleaflet mixing and coupling in liquid-disordered phospholipid bilayers. |
| 26615919 | Biochim. Biophys. Acta | 2016 | Drabik D et al. | The modified fluorescence based vesicle fluctuation spectroscopy technique for determination of lipid bilayer bending properties. |
| 26325346 | Biochim. Biophys. Acta | 2015 | Bavdek A et al. | Enzyme-coupled assays for flip-flop of acyl-Coenzyme A in liposomes. |
| 25450348 | Biochim. Biophys. Acta | 2015 | Kulig W et al. | Experimental determination and computational interpretation of biophysical properties of lipid bilayers enriched by cholesteryl hemisuccinate. |
| 25724816 | Biochim. Biophys. Acta | 2015 | Rui L et al. | Reduced graphene oxide directed self-assembly of phospholipid monolayers in liquid and gel phases. |
| 27806910 | Biochim. Biophys. Acta | 2017 | McDonald C et al. | Structure and function of PspA and Vipp1 N-terminal peptides: Insights into the membrane stress sensing and mitigation. |
| 27480806 | Biochim. Biophys. Acta | 2016 | Freudenthal O et al. | Nanoscale investigation of the interaction of colistin with model phospholipid membranes by Langmuir technique, and combined infrared and force spectroscopies. |
| 27085977 | Biochim. Biophys. Acta | 2016 | Shinoda W | Permeability across lipid membranes. |
| 26657529 | Biochim. Biophys. Acta | 2016 | Chulkov EG and Ostroumova OS | Phloretin modulates the rate of channel formation by polyenes. |
| 25660752 | Biochim. Biophys. Acta | 2015 | Balleza D et al. | Effects of neurosteroids on a model membrane including cholesterol: A micropipette aspiration study. |
| 25542782 | Biochim. Biophys. Acta | 2015 | Lhor M et al. | Structure of the N-terminal segment of human retinol dehydrogenase 11 and its preferential lipid binding using model membranes. |
| 25528473 | Biochim. Biophys. Acta | 2015 | Bulacu M and Sevink GJ | Computational insight in the role of fusogenic lipopeptides at the onset of liposome fusion. |
| 25445675 | Biochim. Biophys. Acta | 2015 | Sakamoto S et al. | Effect of glycyrrhetinic acid on lipid raft model at the air/water interface. |
| 25445669 | Biochim. Biophys. Acta | 2015 | Jobin ML et al. | The role of tryptophans on the cellular uptake and membrane interaction of arginine-rich cell penetrating peptides. |
| 25306964 | Biochim. Biophys. Acta | 2015 | Tavano R et al. | The peculiar N- and (-termini of trichogin GA IV are needed for membrane interaction and human cell death induction at doses lacking antibiotic activity. |
| 25285435 | Biochim. Biophys. Acta | 2015 | Liu Y and Mark Worden R | Size dependent disruption of tethered lipid bilayers by functionalized polystyrene nanoparticles. |
| 25223717 | Biochim. Biophys. Acta | 2015 | Chulkov EG et al. | Membrane dipole modifiers modulate single-length nystatin channels via reducing elastic stress in the vicinity of the lipid mouth of a pore. |