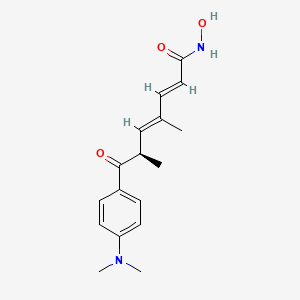
trichostatin A
Trichostatin is a lipid of Polyketides (PK) class. Trichostatin is associated with abnormalities such as Dentatorubral-Pallidoluysian Atrophy, PARAGANGLIOMAS 3, abnormal fragmented structure, Disintegration (morphologic abnormality) and Hyperostosis, Diffuse Idiopathic Skeletal. The involved functions are known as Acetylation, Cell Differentiation process, histone modification, Gene Silencing and Transcriptional Activation. Trichostatin often locates in CD41a, Hematopoietic System, Chromatin Structure, Blood and Endothelium. The associated genes with Trichostatin are SPI1 gene, CELL Gene, Chromatin, CXCR4 gene and DNMT1 gene. The related lipids are Butyrates, Promega, butyrate, Lipopolysaccharides and Steroids. The related experimental models are Knock-out, Mouse Model, Xenograft Model and Cancer Model.
References related to genes published in Mol. Cell. Biol.
| PMID | Journal | Published Date | Author | Title |
|---|---|---|---|---|
| 9774645 | Mol. Cell. Biol. | 1998 | Madisen L et al. | The immunoglobulin heavy chain locus control region increases histone acetylation along linked c-myc genes. |
| 10207061 | Mol. Cell. Biol. | 1999 | Johnson BS et al. | Retinoid X receptor (RXR) agonist-induced activation of dominant-negative RXR-retinoic acid receptor alpha403 heterodimers is developmentally regulated during myeloid differentiation. |
| 10022921 | Mol. Cell. Biol. | 1999 | Dang VD et al. | A new member of the Sin3 family of corepressors is essential for cell viability and required for retroelement propagation in fission yeast. |
| 11158299 | Mol. Cell. Biol. | 2001 | Chang YC et al. | Cooperation of E2F-p130 and Sp1-pRb complexes in repression of the Chinese hamster dhfr gene. |
| 11533236 | Mol. Cell. Biol. | 2001 | Amann JM et al. | ETO, a target of t(8;21) in acute leukemia, makes distinct contacts with multiple histone deacetylases and binds mSin3A through its oligomerization domain. |
| 11340160 | Mol. Cell. Biol. | 2001 | Viollet B et al. | Embryonic but not postnatal reexpression of hepatocyte nuclear factor 1alpha (HNF1alpha) can reactivate the silent phenylalanine hydroxylase gene in HNF1alpha-deficient hepatocytes. |
| 12138181 | Mol. Cell. Biol. | 2002 | Li J et al. | Involvement of histone methylation and phosphorylation in regulation of transcription by thyroid hormone receptor. |
| 11971970 | Mol. Cell. Biol. | 2002 | Fu M et al. | Androgen receptor acetylation governs trans activation and MEKK1-induced apoptosis without affecting in vitro sumoylation and trans-repression function. |
| 11940673 | Mol. Cell. Biol. | 2002 | Curradi M et al. | Molecular mechanisms of gene silencing mediated by DNA methylation. |
| 12391164 | Mol. Cell. Biol. | 2002 | Westendorf JJ et al. | Runx2 (Cbfa1, AML-3) interacts with histone deacetylase 6 and represses the p21(CIP1/WAF1) promoter. |
| 12972616 | Mol. Cell. Biol. | 2003 | Schuettengruber B et al. | Autoregulation of mouse histone deacetylase 1 expression. |
| 12697811 | Mol. Cell. Biol. | 2003 | Zika E et al. | Histone deacetylase 1/mSin3A disrupts gamma interferon-induced CIITA function and major histocompatibility complex class II enhanceosome formation. |
| 12724425 | Mol. Cell. Biol. | 2003 | Jedrusik MA and Schulze E | Telomeric position effect variegation in Saccharomyces cerevisiae by Caenorhabditis elegans linker histones suggests a mechanistic connection between germ line and telomeric silencing. |
| 12861021 | Mol. Cell. Biol. | 2003 | Pivot-Pajot C et al. | Acetylation-dependent chromatin reorganization by BRDT, a testis-specific bromodomain-containing protein. |
| 11113204 | Mol. Cell. Biol. | 2001 | Feng YQ et al. | Position effects are influenced by the orientation of a transgene with respect to flanking chromatin. |
| 15226430 | Mol. Cell. Biol. | 2004 | Naruse Y et al. | Circadian and light-induced transcription of clock gene Per1 depends on histone acetylation and deacetylation. |
| 21791605 | Mol. Cell. Biol. | 2011 | Arzenani MK et al. | Genomic DNA hypomethylation by histone deacetylase inhibition implicates DNMT1 nuclear dynamics. |
| 17353271 | Mol. Cell. Biol. | 2007 | Barr H et al. | Mbd2 contributes to DNA methylation-directed repression of the Xist gene. |
| 15601862 | Mol. Cell. Biol. | 2005 | Goeman F et al. | Growth inhibition by the tumor suppressor p33ING1 in immortalized and primary cells: involvement of two silencing domains and effect of Ras. |
| 16354677 | Mol. Cell. Biol. | 2006 | Solomon JM et al. | Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. |
| 16166624 | Mol. Cell. Biol. | 2005 | Chang CW et al. | Stimulation of GCMa transcriptional activity by cyclic AMP/protein kinase A signaling is attributed to CBP-mediated acetylation of GCMa. |
| 15713621 | Mol. Cell. Biol. | 2005 | Duan H et al. | Histone deacetylase inhibitors down-regulate bcl-2 expression and induce apoptosis in t(14;18) lymphomas. |
| 16479005 | Mol. Cell. Biol. | 2006 | Pedram M et al. | Telomere position effect and silencing of transgenes near telomeres in the mouse. |
| 17145766 | Mol. Cell. Biol. | 2007 | Fang S et al. | Coordinated recruitment of histone methyltransferase G9a and other chromatin-modifying enzymes in SHP-mediated regulation of hepatic bile acid metabolism. |
| 17101789 | Mol. Cell. Biol. | 2007 | Zhang R et al. | HP1 proteins are essential for a dynamic nuclear response that rescues the function of perturbed heterochromatin in primary human cells. |
| 18541673 | Mol. Cell. Biol. | 2008 | Lecona E et al. | Upregulation of annexin A1 expression by butyrate in human colon adenocarcinoma cells: role of p53, NF-Y, and p38 mitogen-activated protein kinase. |
| 18710935 | Mol. Cell. Biol. | 2008 | Obrdlik A et al. | The histone acetyltransferase PCAF associates with actin and hnRNP U for RNA polymerase II transcription. |